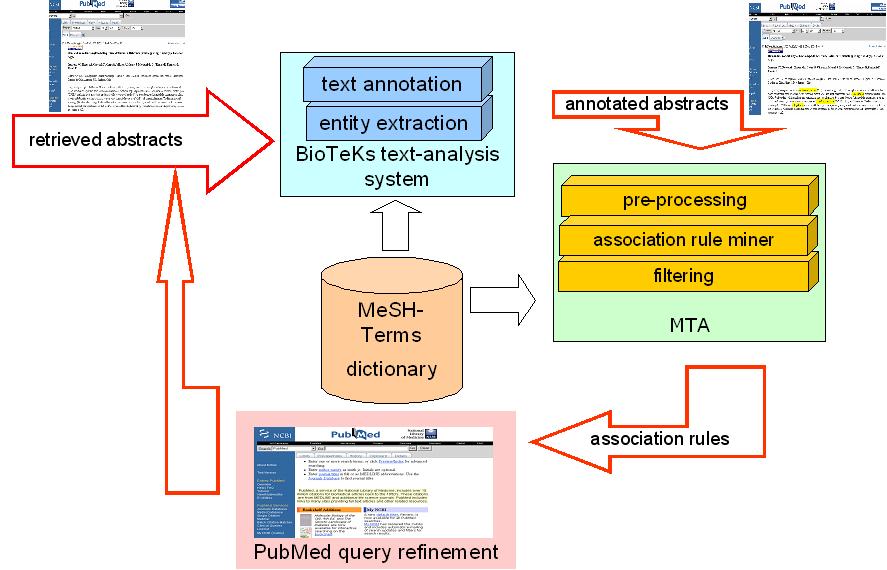
A framework for MTA in PubMed query expansion tasks.
|
|
| A short description
The functional architecture The distribution package Project team Related publications |
 |
|
A framework for MTA in PubMed query expansion tasks. |
Warning: The system MTA is free for evaluation, research and teaching purposes, but not for commercial purposes.
Please Acknowledge